| study | p_1 | p_2 | q_1 | q_2 | n |
|---|---|---|---|---|---|
| 1 | 0.4830 | 0.3920 | 0.4610 | 0.4160 | 2046 |
| 2 | 0.4100 | 0.3500 | 0.4200 | 0.3600 | 1149 |
| 3 | 0.4660 | 0.3310 | 0.4650 | 0.3490 | 1112 |
| 4 | 0.4601 | 0.3222 | 0.4082 | 0.3586 | 1112 |
| 5 | 0.4400 | 0.3200 | 0.3970 | 0.3300 | 1082 |
| 6 | 0.3880 | 0.2930 | 0.3820 | 0.3060 | 1484 |
An example survey
An example survey
\[ \newcommand{\mc}[1]{\mathcal{#1}} \newcommand{\R}{\mathbb{R}} \newcommand{\E}{\mathbb{E}} \renewcommand{\P}{\mathbb{P}} \newcommand{\var}{{\rm Var}} % Variance \newcommand{\mse}{{\rm MSE}} % MSE \newcommand{\bias}{{\rm Bias}} % MSE \newcommand{\cov}{{\rm Cov}} % Covariance \newcommand{\iid}{\stackrel{\rm iid}{\sim}} \newcommand{\ind}{\stackrel{\rm ind}{\sim}} \renewcommand{\choose}[2]{\binom{#1}{#2}} % Choose \newcommand{\chooses}[2]{{}_{#1}C_{#2}} % Small choose \newcommand{\cd}{\stackrel{d}{\rightarrow}} \newcommand{\cas}{\stackrel{a.s.}{\rightarrow}} \newcommand{\cp}{\stackrel{p}{\rightarrow}} \newcommand{\bin}{{\rm Bin}} \newcommand{\ber}{{\rm Ber}} \DeclareMathOperator*{\argmax}{argmax} \DeclareMathOperator*{\argmin}{argmin} \]
- There are 6 surveys and each survey has two questions:
- prefer A or B, or neither?
- prefer A’ or B, or neither?
- Notations:
- \(\hat{p}_1\): proportion of people who prefer A over B.
- \(\hat{p}_2\): proportion of people who prefer B over A.
- \(\hat{q}_1\): proportion of people who prefer A’ over B.
- \(\hat{q}_2\): proportion of people who prefer B over A’.
- \(n\) is the sample size in each survey
Survey Results
- Two possible statistics:
- difference in proportion: \(T_1 = \hat{p}_1 - \hat{q}_1\).
- difference in difference: \(T_2 = (\hat{p}_1 - \hat{p}_2) - (\hat{q}_1 - \hat{q}_2)\)
Assumptions
- The surveys are independent.
- The responses to the two questions in each survey are also independent.
- The surveys are conducted based on simple random samples.
Parametric Bootstrap
B <- 10^4
T1_boot <- matrix(0, ncol = B, nrow = 6)
T2_boot <- matrix(0, ncol = B, nrow = 6)
set.seed(2023)
for (i in 1:B) {
for (j in 1:6) {
prop_boot_p <- rmultinom(1, n[j],
prob = c(p_1[j], p_2[j], 1-p_1[j]-p_2[j]))/n[j]
prop_boot_q <- rmultinom(1, n[j],
prob = c(q_1[j], q_2[j], 1-q_1[j]-q_2[j]))/n[j]
T1_boot[j, i] <- prop_boot_p[1] - prop_boot_q[1]
T2_boot[j, i] <- (prop_boot_p[1] - prop_boot_p[2]) -
(prop_boot_q[1] - prop_boot_q[2])
}
}Difference in Proportions
survey_dp_boot <- tibble(study = 1:6,
DiP = 100 * rowMeans(T1_boot),
se = 100 * apply(T1_boot, 1, sd),
CI.lower = 100 * apply(T1_boot, 1, quantile,
probs = 0.025),
CI.upper = 100 * apply(T1_boot, 1, quantile,
probs = 0.975))
survey_dp_boot |> kable(digits = 2)| study | DiP | se | CI.lower | CI.upper |
|---|---|---|---|---|
| 1 | 2.18 | 1.54 | -0.83 | 5.23 |
| 2 | -0.97 | 2.05 | -4.96 | 3.05 |
| 3 | 0.11 | 2.12 | -4.05 | 4.23 |
| 4 | 5.14 | 2.09 | 0.99 | 9.26 |
| 5 | 4.30 | 2.14 | 0.09 | 8.50 |
| 6 | 0.63 | 1.79 | -2.90 | 4.11 |
Difference in Differences
survey_did_boot <- tibble(study = 1:6,
DiD = 100 * rowMeans(T2_boot),
se = 100 * apply(T2_boot, 1, sd),
CI.lower = 100 * apply(T2_boot, 1, quantile,
probs = 0.025),
CI.upper = 100 * apply(T2_boot, 1, quantile,
probs = 0.975))
survey_did_boot |> kable(digits = 2)| study | DiD | se | CI.lower | CI.upper |
|---|---|---|---|---|
| 1 | 4.57 | 2.88 | -1.03 | 10.22 |
| 2 | 0.05 | 3.63 | -6.96 | 7.14 |
| 3 | 1.89 | 3.78 | -5.58 | 9.35 |
| 4 | 8.74 | 3.69 | 1.44 | 15.92 |
| 5 | 5.29 | 3.70 | -1.85 | 12.66 |
| 6 | 1.95 | 3.03 | -4.04 | 7.88 |
Bayesian Analysis
Joint distribution of the two questions
Apparently, the responses to the two question are correlated.
However we only the marginal distributions for each question.
We need to model the joint distribution directly.
Let Q1 be the first question (A vs B) and Q2 be the second question (A’ vs B).
Suppose the joint distribution is
Q1\Q2 A’ B no response A \(\rho_1\) \(\rho_{12}\) \(\gamma\) B \(\rho_{21}\) \(\rho_2\) \(\gamma\) no response \(\gamma\) \(\gamma\) \(\rho_3\) The parameter is \(\theta = (\rho_1, \rho_2, \rho_3, \rho_{12}, \rho_{21}, \gamma)\), where \(\rho_1 + \rho_2 + \rho_3 + \rho_{12} + \rho_{21} + 4\gamma = 1\).
Denote the counts by \(n_{ij}\), where \(i,j \in \{1,2,3\} = \{A, B, \text{no response}\}\).
Interpretation of the parameters
- \(\rho_1\): the proportion of people that always prefer A type.
- \(\rho_2\): the proportion of people that always prefer B type.
- \(\rho_3\): the proportion of people that always give no response.
- \(\rho_{12}\): the proportion of people that prefer \(A > B > A'\).
- \(\rho_{21}\): the proportion of people that prefer \(A' > B > A\).
- \(\gamma\): the proportion of people that answer only one of the two questions.
Missing Data
- We have observed \(Y = [n_{1\cdot}, n_{2\cdot}, n_{3\cdot},n_{\cdot 1}, n_{\cdot 2}]^T\), where \(n_{i\cdot} = \sum_j n_{ij}\) and \(n_{\cdot j} = \sum_i n_{ij}\).
- Let \(U = [n_{11}, n_{12}, n_{13}, n_{21}, n_{22}, n_{23}, n_{31}, n_{32}, n_{33}]^T\) be the complete data.
- If we also observe \(Z = [n_{11}, n_{12}, n_{21}, n_{32}]^T\), then \(U\) can be recovered from \(Y\) and \(Z\).
- \(Z\) is called the missing data.
- Let \(M\) be a matrix such that \[ U = M\left[\begin{array}{c} Y \\ Z \end{array}\right]. \]
- The distribution of \(U\) multinomial with parameter \((\rho_1, \rho_{12}, \gamma, \rho_{21}, \rho_2, \gamma, \gamma, \gamma, \rho_3)\).
Intuition
- If we observe \(U\), we can find the posterior \(\pi(\theta \mid U)\).
- Observing \(U\) is equivalent to observing \(Y\) and \(Z\) and hence the posterior can be written as \(\pi(\theta \mid Y, Z)\).
- However, \(Z\) is missing, so we can not condition on \(Z\).
- The most we can do is \(\pi(\theta, Z \mid Y)\).
- By Bayes Theorem, \[ \pi(\theta, Z \mid Y) = \frac{p(Y, Z \mid \theta)\pi(\theta)}{p(Y)}. \]
- The distribution \(p(Y, Z \mid \theta)\) is available since \([Y^T, Z^T]^T = M^{-1}U\) and \(U\) is multinomial.
- We can also consider the marginal posterior \(\pi(\theta \mid Y) = \sum_{Z} \pi(\theta, Z \mid Y)\).
Data (Survey 1)
| Q1\Q2 | A’ | B | No response | Total |
|---|---|---|---|---|
| A | \(\textcolor{red}{437}\) | \(\textcolor{red}{409}\) | \(\textcolor{red}{120}\) | 988 |
| B | \(\textcolor{red}{358}\) | \(\textcolor{red}{327}\) | \(\textcolor{red}{99}\) | 802 |
| No response | \(\textcolor{red}{127}\) | \(\textcolor{red}{96}\) | \(\textcolor{red}{27}\) | 256 |
| Total | 943 | 851 | \(\textcolor{blue}{252}\) | \(\textcolor{blue}{2046}\) |
- The numbers in black are observed.
- The numbers in red are my random guesses.
- The numbers in blue are redundant as they can be computed from the others.
Sampling Algorithm
- We need to sample from \(\pi(\theta, Z \mid Y)\).
- The sampling is achieved by Gibbs sampler:
- sample \(Z\) from \(\pi(Z \mid \theta, Y)\)
- sample \(\theta\) from \(\pi(\theta \mid Z, Y)\).
- In our case, \(\pi(Z \mid \theta, Y)\) is independent of \(\theta\) and the full conditionals \(\pi(z_i \mid z_{-i}, Y)\) are hypergeometric distributions.
- You fill find that the derivation is the same as Fisher’s exact test.
- The distribution \(\pi(\theta \mid Z, Y)\) is exactly \(\pi(\theta \mid U)\), i.e., the posterior given the complete data.
Prior and posterior
- We need to specify a prior for \(\theta = (\rho_1, \rho_2, \rho_3, \rho_{12}, \rho_{21}, 4\gamma)\).
- For convenience, we choose \((\rho_1, \rho_2, \rho_3, \rho_{12}, \rho_{21}, 4\gamma) \sim \text{Dir}(\alpha_1, \ldots, \alpha_6)\).
- The posterior given the complete data is \[\begin{multline*} (\rho_1, \rho_2, \rho_3, \rho_{12}, \rho_{21}, 4\gamma) \mid U \sim \text{Dir}(\alpha_1 + n_{11}, \alpha_2 + n_{22}, \alpha_3 + n_{33}, \alpha_4 + n_{12},\\ \alpha_5 + n_{21}, \alpha_6 + n_{13} + n_{23} + n_{31} + n_{32}). \end{multline*}\]
Analysis for Survey 1
| Q_2.A' | Q_2.B | Q_2.No response | total | |
|---|---|---|---|---|
| Q_1.A | 22.23 | 20.07 | 5.43 | 47.73 |
| Q_1.B | 18.09 | 16.30 | 5.43 | 39.82 |
| Q_1.No response | 5.43 | 5.43 | 1.59 | 12.45 |
| total | 45.75 | 41.80 | 12.45 | 100.00 |
Analysis for Survey 2
| Q_2.A' | Q_2.B | Q_2.No response | total | |
|---|---|---|---|---|
| Q_1.A | 17.19 | 14.81 | 8.83 | 40.83 |
| Q_1.B | 14.75 | 12.58 | 8.83 | 36.17 |
| Q_1.No response | 8.83 | 8.83 | 5.33 | 23.00 |
| total | 40.77 | 36.23 | 23.00 | 100.00 |
Meta analysis
- We can also perform meta analysis by combining the 6 surveys.
- Let \(U_i\) be the complete data for survey \(i\).
- A simple hierarchical model is \[\begin{align*} U_i \mid \theta_i &\ind \text{Multinomial}(\theta_i),\\ \theta_i \mid \alpha, \nu &\iid \text{Dir}(\alpha\nu),\\ \nu &\sim \text{Dir}(1, 1, 1, 1, 1, 1),\\ \alpha &\sim \text{Exp}(0.001) \end{align*}\]
- The complete data \(U_i\) is obtained by generating the missing data \(Z_i\) from \(\pi(Z_i \mid Y_i)\) and then combining with the observed data \(Y_i\).
- I wrote a function
multi_survey_samplerto perform the meta analysis.
What is the question?
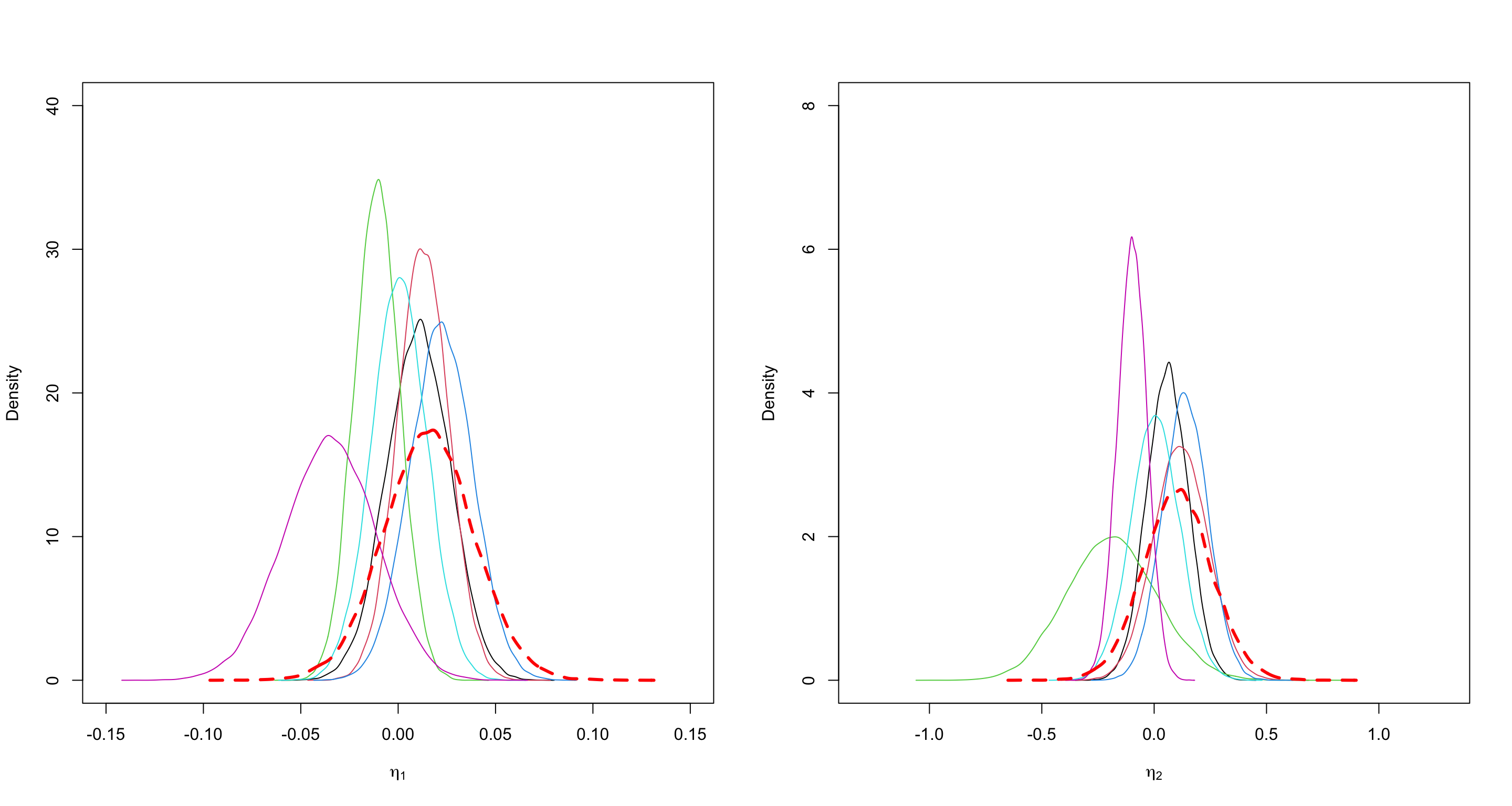
- Suppose you have obtained posterior samples of \(\theta_i = (\rho_{1,i}, \rho_{2,i}, \rho_{3,i}, \rho_{12,i}, \rho_{21,i}, \gamma_i)\) for \(i = 1, \ldots, 6\).
- What do you want to know?
- Probably, you want to compare \(\rho_{12,i}\) and \(\rho_{21,i}\) for each \(i = 1, \ldots, 6\).
- You have several choices:
- use \(\eta_{1, i} = \rho_{12,i} - \rho_{21,i} \in [-1, 1]\);
- use \(\eta_{2, i} = \log\frac{\rho_{12,i}}{\rho_{21,i}} \in (-\infty, \infty)\);
- Since we are using the joint distribution, it is okay to subtract probabilities (although it is not recommended).
- You cannot arbitrarily subtract conditional probabilities or marginal probabilities.
- For example, it is meaningless to compute \[\begin{align*} & \P(X = 1) - \P(Y = 0) \quad \text{or}\\ & \P(X = 1 \mid Y = 1) - \P(X = 1 \mid Y = 0). \end{align*}\]
Analysis
source("../dataset/survey/survey_Missing.R")
survey_data <- read.csv("../dataset/survey/survey_data.csv",header = TRUE)
attach(survey_data)
N1 <- cbind(round(n*p_1), round(n*p_2))
N1 <- cbind(N1, n - N1[,1] - N1[,2])
N2 <- cbind(round(n*q_1), round(n*q_2))
N2 <- cbind(N2, n - N2[,1] - N2[,2])
detach(survey_data)
Y <- cbind(N1, N2[,1:2])
Y |> `colnames<-`(c("n_1.", "n_2.", "n_3.", "n_.1", "n_.2")) |> kable()| n_1. | n_2. | n_3. | n_.1 | n_.2 |
|---|---|---|---|---|
| 988 | 802 | 256 | 943 | 851 |
| 471 | 402 | 276 | 483 | 414 |
| 518 | 368 | 226 | 517 | 388 |
| 512 | 358 | 242 | 454 | 399 |
| 476 | 346 | 260 | 430 | 357 |
| 576 | 435 | 473 | 567 | 454 |
Analysis
Results
- \(\eta_1 = \rho_{12} - \rho_{21} =\) (prefer A > B > A’) - (prefer A’ > B > A)
- \(\eta_2 = \log\frac{\rho_{12}}{\rho_{21}} =\) log(prefer A > B > A’) - log(prefer A’ > B > A)
- The red dashed line is the overall estimate.